Gateway User Guide
This Guide will guide the user through four in silico platforms: VEGAHUB, OCHEM, the Danish (Q)SAR Database and AMBIT in order to help how the platforms work.
GUIDE TO THE USE OF THE GATEWAY
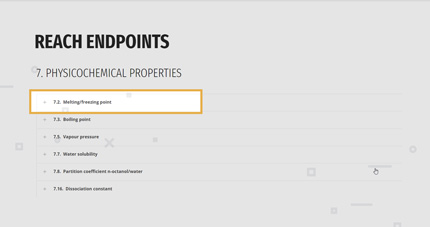
1. REACH ENDPOINTS
According to his/her needs, the user can filter the models by the endpoints list, as in the REACH regulation.
The main categories are:
- PHYSICOCHEMICAL PROPERTIES
- TOXICOLOGICAL INFORMATION
- ECOTOXICOLOGICAL INFORMATION
In addition to the REACH’s categories, the gateway reports the models also for ENDOCRINE ACTIVITY
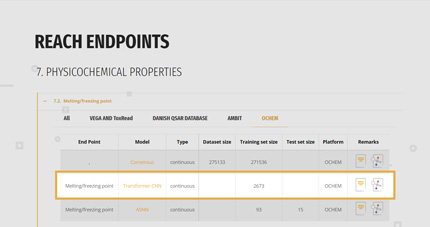
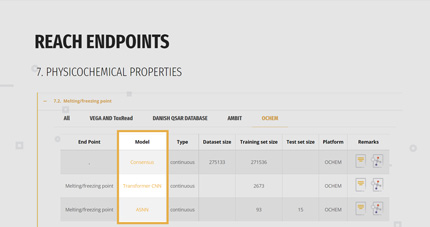
2. SELECTION OF THE SUITABLE MODEL
For each endpoint, the gateway reports a list of the available models with their basilar information:
- Name of the model,
- If the prediction will be a classification or a continuous value,
- Size of the dataset at the base of the model, comprising the splitting in training and test set if available,
- Direct link to the model/platform and to the model’s documentation like QMRF and papers.
The user can look at the whole list or he/she can select only those models belonging to a specific platform.
3. PREDICTING
Once selected the model of interest, click on the link present in the “Model” column; you will be redirected to the access page of the models.
Each platform works differently:
VEGA
From the VEGA web site, it will be possible to freely downloading the stand-alone application, and easily use it thank to an intuitive guide.
VEGA has about 100 freely available (Q)SAR models covering most of the REACH endpoints.
Danish (Q)SAR Database

Clicking on “Enter” it will be possible to explore the estimates for more than 600,000 chemical substances, from free and commercial platforms, or to directly predict your own molecule by choosing the “Danish (Q)SAR models” option
OCHEM
Once logged in, it is possible to access OCHEM platform and take advantage of the various (Q)SAR models and databases within it.
It is also possible developing his own model.
AMBIT
From the linked page it is possible to access the description of the tools available in AMBIT and the various access methods
4. INTERPRETATION OF THE RESULTS
The user can consult all the available documentation of the in silico tools in the dedicated section.
Here an example about the integration of the results from multiple models
